Nanonet Force Microscopy

At STEP lab, we have devised a suspended fiber-based force measurement platform (Nanonet Force Microscopy, NFM) using ECM-mimicking anisotropic fibers. NFM estimates cell forces from deflection of fibers (inwards or outwards) as cells tug or push on them. We deposit large diameter fibers (μm) that act as supporting structures (‘base fibers’). Smaller diameter fibers are deposited orthogonal to base fibers at desired spacing and fused at the intersections to achieve fixed-fixed boundary conditions. Our method establishes force vectors that originate from the focal adhesion sites and are directed along the major actin stress fibers (force bearing elements). A combination of beam mechanics and optimization techniques allows us to estimate forces by minimizing the error between finite element model predictions and measured experimental fiber deflections. NFM can be used to study the two-broad classification of forces: innate contractility inside-out (IO) forces and externally applied forces outside-In (OI). The unique fiber based platform has allowed us to demonstrate the following specific application
Representative Publications
Exogenous high-voltage pulses increase cell membrane permeability through a phenomenon known as electroporation. This process may also disrupt the cell cytoskeleton causing changes in cell contractility; however, the contractile signature of cell force after electroporation remains unknown. Here, single-cell forces post-electroporation are measured using suspended extracellular matrix-mimicking nanofibers that act as force sensors. Ten, 100 μs pulses are delivered at three voltage magnitudes (500, 1000, and 1500 V) and two directions (parallel and perpendicular to cell orientation), exposing glioblastoma cells to electric fields between 441 V cm–1 and 1366 V cm–1. Cytoskeletal-driven force loss and recovery post-electroporation involves three distinct stages. Low electric field magnitudes do not cause disruption, but higher fields nearly eliminate contractility 2–10 min post-electroporation as cells round following calcium-mediated retraction (stage 1). Following rounding, a majority of analyzed cells enter an unusual and unexpected biphasic stage (stage 2) characterized by increased contractility tens of minutes post-electroporation, followed by force relaxation. The biphasic stage is concurrent with actin disruption-driven blebbing. Finally, cells elongate and regain their pre-electroporation morphology and contractility in 1–3 h (stage 3). With increasing voltages applied perpendicular to cell orientation, we observe a significant drop in cell viability. Experiments with multiple healthy and cancerous cell lines demonstrate that contractile force is a more dynamic and sensitive metric than cell shape to electroporation. A mechanobiological understanding of cell contractility post-electroporation will deepen our understanding of the mechanisms that drive recovery and may have implications for molecular medicine, genetic engineering, and cellular biophysics.
Phillip Graybeal and Aniket Jana
Co-developed with Rafael Davalos
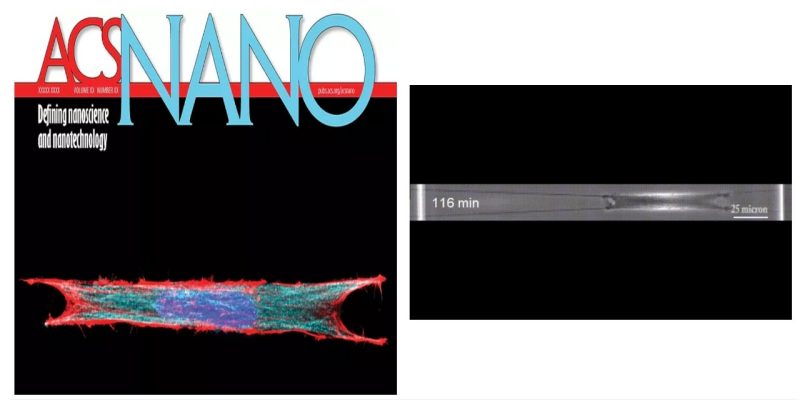
Aligned extracellular matrix fibers enable fibroblasts to undergo myofibroblastic activation and achieve elongated shapes. Activated fibroblasts adhere to the extracellular fibers and contract, perpetuating the alignment of these fibers. This poorly understood feedback process is critical in chronic fibrosis conditions, including cancer. Here, using fiber networks that serve as force sensors, we identify “3D perpendicular lateral protrusions” (3D-PLPs) that evolve from lateral cell extensions named twines. The specific morphology of PLPs enables them to exert force on parallel neighboring fibers, causing cells to increase their contractility. Twines originate from stratification of cyclic actin-waves traversing the cell and swing freely in 3D to engage neighboring fibers. Once engaged, a lamellum forms and extends multiple secondary twines, which fill in to form a sheet-like PLP, in a force- entailing process that transitions focal adhesions to elongated 3D-adhesions. Controlling the geometry of extracellular networks confirms that anisotropic fibrous environments support 3D-PLP formation and function, suggesting an explanation for cancer-associated desmoplastic expansion at single-cell resolution.
Abinash Padhi
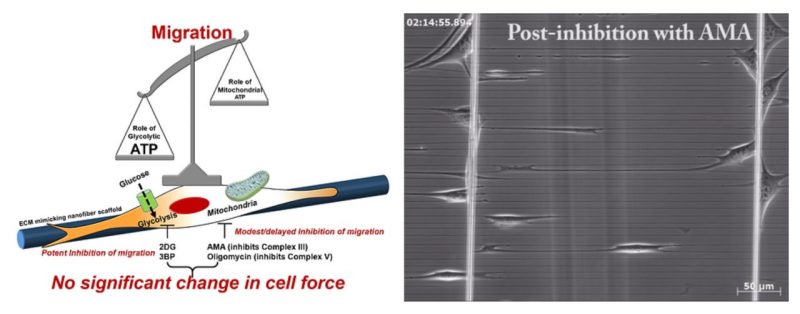
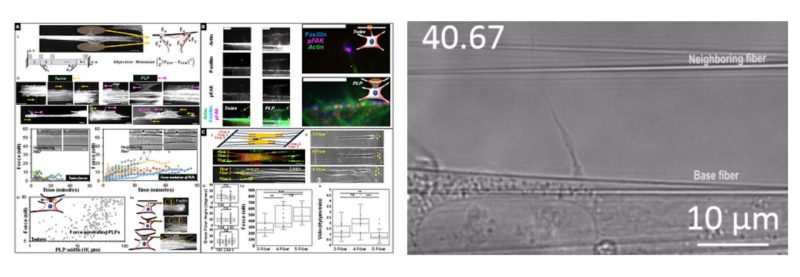
Cell migration is centrally involved in a myriad of physiological processes, including morphogenesis, wound healing, tissue repair, and metastatic growth. The bioenergetics that underlie migratory behavior are not fully understood, in part because of variations in cell culture media and utilization of experimental cell culture systems that do not model physiological connective extracellular fibrous networks. In this study, we evaluated the bioenergetics of C2C12 myoblast migration and force production on fibronectin-coated nanofiber scaffolds of controlled diameter and alignment, fabricated using a nonelectrospinning spinneret-based tunable engineered parameters (STEP) platform. The contribution of various metabolic pathways to cellular migration was determined using inhibitors of cellular respiration, ATP synthesis, glycolysis, or glucose uptake. Despite immediate effects on oxygen consumption, mitochondrial inhibition only modestly reduced cell migration velocity, whereas inhibitors of glycolysis and cellular glucose uptake led to striking decreases in migration. The migratory metabolic sensitivity was modifiable based on the substrates present in cell culture media. Cells cultured in galactose (instead of glucose) showed substantial migratory sensitivity to mitochondrial inhibition. We used nanonet force microscopy to determine the bioenergetic factors responsible for single-cell force production and observed that neither mitochondrial nor glycolytic inhibition altered single-cell force production. These data suggest that myoblast migration is heavily reliant on glycolysis in cells grown in conventional media. These studies have wide-ranging implications for the causes, consequences, and putative therapeutic treatments aimed at cellular migration.
- Abinash Padhi
- Selected as APS Select and nominated for best graduate student paper award 2019
- Study led by Brown Lab, Virginia Tech
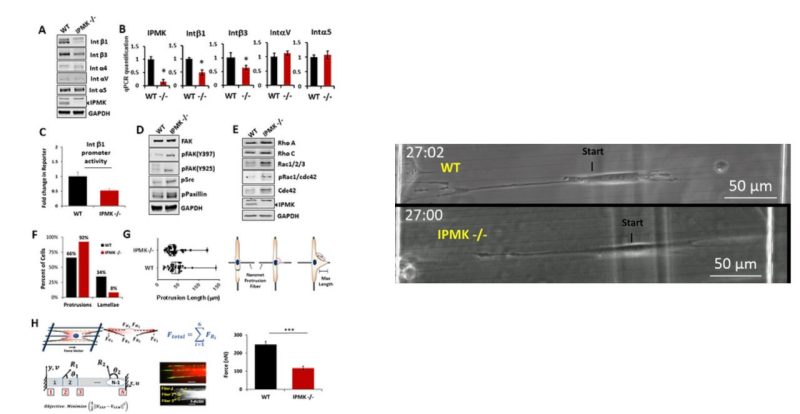
Metformin has been shown to alter cell adhesion protein expression, which is thought to play a role in its observed antitumor properties. We found that metformin treatment down‐regulated integrin β1 concomitant with the loss of inositol polyphosphate multikinase (IPMK) in murine myocytes, adipocytes, and hepatocytes. To determine if IPMK was upstream of integrin β1 expression, we examined IPMK‐/‐ mouse embryonic fibroblast cells and found that integrins β1and β3 gene expression was reduced by half, relative to wild‐type cells, whereas focal adhesion kinase (FAK) activity and Rho/Rac/Cdc42 protein levels were increased, resulting in migration defects. Using nanonet force microscopy, we determined that cell:extracellular matrix adhesion and cell contractility forces were decreased, confirming the functional relevance of integrin and Rho protein dysregulation. Pharmacological studies showed that inhibition of both FAK1 and proline‐rich tyrosine kinase 2 partially restored integrin β1 expression, suggesting negative regulation of integrin β1 by FAK. Together our data indicate that IPMK participates in the regulation of cell migration and provides a potential link between metformin and wound healing impairment.
Abinash Padhi, study led by Kim Lab, Johns Hopkins University
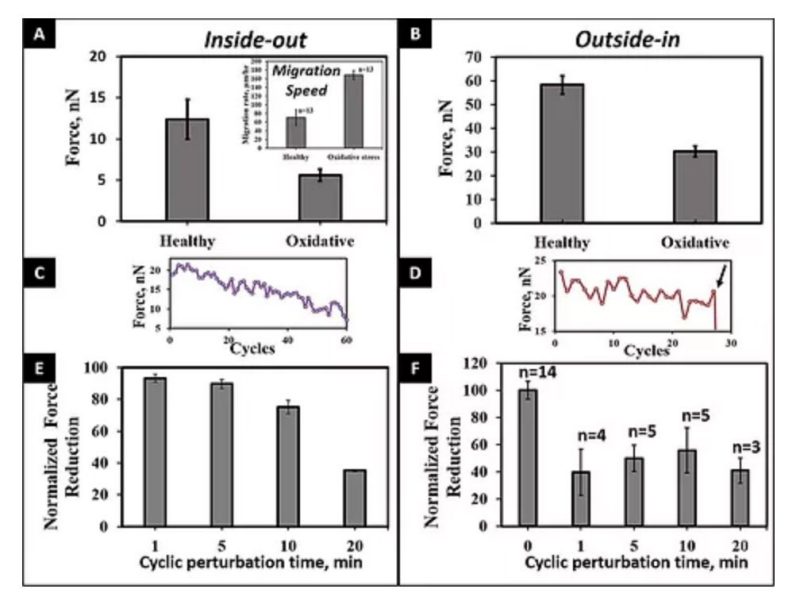
A number of innovative methods exist to measure cell–matrix adhesive forces, but they have yet to accurately describe and quantify the intricate interplay of a cell and its fibrous extracellular matrix (ECM). In cardiovascular pathologies, such as aortic aneurysm, new knowledge on the involvement of cell–matrix forces could lead to elucidation of disease mechanisms. To better understand this dynamics, we measured primary human aortic single smooth muscle cell (SMC) forces using nanonet force microscopy in both inside-out (I-O intrinsic contractility) and outside-in (O-I external perturbation) modes. For SMC populations, we measured the I-O and O-I forces to be 12.9 ± 1.0 and 57.9 ± 2.5 nN, respectively. Exposure of cells to oxidative stress conditions caused a force decrease of 57 and 48% in I-O and O-I modes, respectively, and an increase in migration rate by 2.5-fold. Finally, in O-I mode, we cyclically perturbed cells at constant strain of varying duration to simulate in vivo conditions of the cardiac cycle and found that I-O forces decrease with increasing duration and O-I forces decreased by half at shorter cycle times. Thus our findings highlight the need to study forces exerted and felt by cells simultaneously to comprehensively understand force modulation in cardiovascular disease.
Alexander Hall
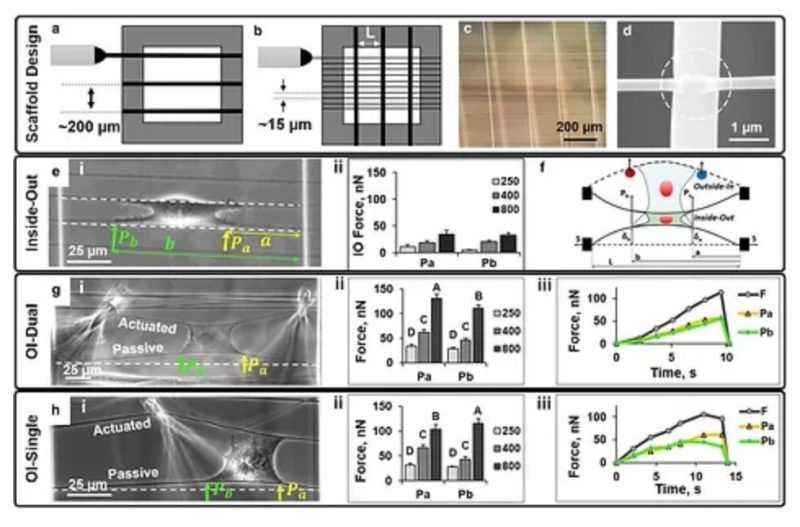
The influence of physical forces exerted by or felt by cells on cell shape, migration, and cytoskeleton arrangement is now widely acknowledged and hypothesized to occur due to modulation of cellular inside-out forces in response to changes in the external fibrous environment (outside-in). Our previous work using the non-electrospinning Spinneret-based Tunable Engineered Parameters’ suspended fibers has revealed that cells are able to sense and respond to changes in fiber curvature and structural stiffness as evidenced by alterations to focal adhesion cluster lengths. Here, we present the development and application of a suspended nanonet platform for measuring C2C12 mouse myoblast forces attached to fibers of three diameters (250, 400, and 800 nm) representing a wide range of structural stiffness (3–50 nN/μm). The nanonet force microscopy platform measures cell adhesion forces in response to symmetric and asymmetric external perturbation in single and cyclic modes. We find that contractility-based, inside-out forces are evenly distributed at the edges of the cell, and that forces are dependent on fiber structural stiffness. Additionally, external perturbation in symmetric and asymmetric modes biases cell-fiber failure location without affecting the outside-in forces of cell-fiber adhesion. We then extend the platform to measure forces of (1) cell-cell junctions, (2) single cells undergoing cyclic perturbation in the presence of drugs, and (3) cancerous single-cells transitioning from a blebbing to a pseudopodial morphology.
Kevin Sheets
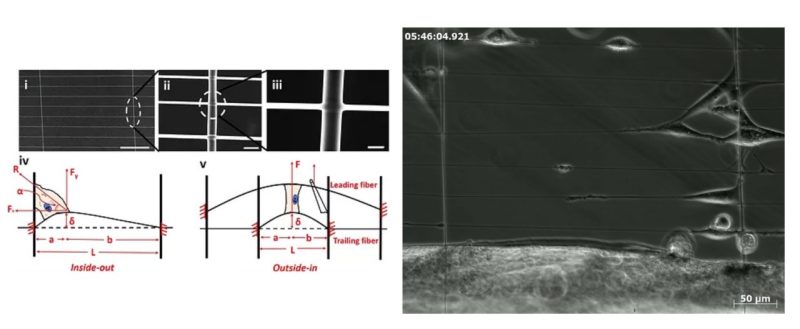
The role of physical forces in disease onset and progression is widely accepted and this knowledge presents an alternative route to investigating disease models. Recently, numerous force measurement techniques have been developed to probe single and multi-cell behavior. While these methods have yielded fundamental insights, they are yet unable to capture the fibrous extra-cellular matrix biophysical interactions, involving parameters of curvature, structural stiffness (N m−1), alignment and hierarchy, which have been shown to play key roles in disease and developmental biology. Using a highly aggressive glioma model (DBTRG-05MG), we present a platform technology to quantify single cell force modulation (both inside-out and outside-in) with and without the presence of a cytoskeleton altering drug (cytochalasin D) using suspended and aligned fiber networks (nanonets) beginning to represent the aligned glioma environment. The nanonets fused in crisscross patterns were manufactured using the non-electrospinning spinneret based tunable engineering parameters technique. We demonstrate the ability to measure contractile single cell forces exerted by glioma cells attached to and migrating along the fiber axis (inside-out). This is followed by a study of force response of glioma cells attached to two parallel fibers using a probe deflecting the leading fiber (outside-in). The forces are calculated using beam deflection within the elastic limit. Our data shows that cytochalasin D compromises the spreading area of single glioma cells, eventually decreasing their 'inside-out' contractile forces, and 'outside-in' force response to external strain. Most notably, for the first time, we demonstrate the feasibility of using physiologically relevant aligned fiber networks as ultra-sensitive force (~nanoNewtons) probes for investigating drug response and efficacy in disease models at the single cell resolution.
Puja Sharma


